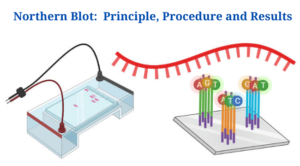
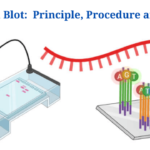
What Is Northern Blot?
For the study of RNA in complicated mixtures, Northern blot is a technique that relies on the notion of “blotting”.
- Essentially, it is a modified version of the Southern Blotting technique, which was originally developed for the examination of DNA sequences.
- Molecular biology requires the detection of certain sequences of nucleic acids that have been isolated from various types of biological samples, which is why blotting techniques are so important in the discipline.
- The idea is the same as southern blotting, except that northern blotting identifies RNA sequences, while southern blotting does not.
- In this way, you may find out how long the RNA sequences are and if there are any differences in them.
- It has also been used to quantify RNA sequences, despite its primary concentration on RNA sequence identification.
- Numerous improvements have been made to the technique since its development, allowing it to be used to analyse mRNAs, pre-mRNAs and short RNAs.
- However, newer and more cost-effective techniques like as RT-PCR have gradually replaced Northern Blotting as the predominant approach for the analysis of RNA fragments.
Created with BioRender
Northern Blot Principle
- No matter what technique you use, all biomolecules must be transferred from one membrane to another in order for a Northern Blot to work.
- Gel electrophoresis is used to separate the RNA samples based on their size. Single-stranded RNAs can form secondary structures through intermolecular base pairing because of their single-stranded nature. Under these conditions, RNA segments are electrophoretically separated.
- A nylon membrane is used to transport the RNA fragments. RNA cannot bind to nitrocellulose membrane, hence it isn’t employed.
- Fixing agents immobilise the transferred segments on the membrane. Addition of tagged probe corresponding to RNA sequences on the membrane detects RNA fragments on the membrane.
- To detect RNA, it is necessary to use hybridization, because it allows for accurate segment identification due to its specificity.
- This technique uses size-dependent separation of RNA segments and may therefore be used to calculate the transcript sizes.
Northern Blot Requirements
Equipment
- Casting with Agarose Gel
- Supply of Electricity
- Microwave
- Centrifuge
- Block of heat
- UV-crosslinking agent (UVC)
- Oven for hybridization
- vessels of hybridization
- Vials
- Forceps
- Pipettes
- Tubes made of glass are also available.
Materials
- The gel of agarose
- Citrate of sodium
- Dehydrate of ethylenediaminetetraacetic acid disodium salt
- NaOH
- HCl
- Formaldehyde
- Glycerol
- Bromure of ethidium
- It’s a blue dye called bromophenol.
- Ladder of RNA
- MgCl2
- NaCl
- Polyvinylpyrrolidone
- Bovine Serum Albumin
- SDS
- NaH2PO4
- Tris-HCl
- Triton-X
- DTT
- Taq buffer
- Taq polymerase.
How to perform a Northern Blot
1 . On a denaturing gel, the separation of RNA
- Formaldehyde is added to the agarose solution to create the RNA gel solution.
- Assemble the cast, and pour in the denaturing gel that you‘ve made earlier in the day. Wells are formed by adding a toothed comb to the gel while it sets.
- At this point you can either remove the comb or let it sit for 30 minutes before running the gel.
- An equal volume of loading buffer and 15 g of RNA are combined together. Add three micrograms of DNA marker to the same amount of loading buffer.
- A heating block at 65°C is used to incubate the samples for roughly 12-15 minutes.
- They’re then added to an equilibrated gel with RNA markers in the first row of wells.
- Next, the gel is run at 125V for approximately three hours.
2. Gel-to-nylon membrane transfer of RNA
- Cut a nylon membrane slightly larger than the denaturing gel. Prepare filter paper with similar size as nylon membrane.
- Electrophoresis is completed by taking out the RNA gel and rinsing it under running water.
- This is done by placing a rectangular sponge somewhat larger than gel on a glass dish, and then filling the glass dish with SSC to a point where the sponge is about half-submerged.
- On top of the sponge, a few pieces of Whatman 3mm paper are wetted with SSC buffer and then placed on top of the sponge.
- To eliminate air bubbles, the gel is rolled over the filter paper with a glass pipette and squeezed.
- The prepared nylon membrane is soaked in distilled water for roughly 5 minutes on an RNase-free dish.
- Wet membrane is placed on top of the gel without allowing air bubbles to form on the surface of the material.
- On top of the membrane, a few more filter papers are added and the surface is again saturated with SSC.
- All of the pieces are held together by an extra-large glass plate that is placed on top of everything. In order to achieve a successful transfer, the structure is left in place over night.
3. Immobility
- Gel is removed and rinsing with SSC is performed when the transfer is complete.
- Filter paper and membrane are sandwiched together in a vacuum oven at 80°C and cooked for 2 hours.
- UV transilluminator irradiates the membrane for an adequate period of time encased in UV transparent plastic wrap.
4. Hybridization
- It is required that DNA or RNA probes be tagged to a particular activity of >108 dpm/g and that unincorporated nucleotides be eliminated.
- SSC is used to moisten the membrane containing the immobilised RNA.
- 1ml of formaldehyde solution is introduced to a hybridization tube with the membrane towards the RNA.
- Hybridization takes place in a chamber at 42°C for three hours.
- After 10 minutes at 100°C in a water bath or incubator, the double-stranded probe will be denatured.
- After pipetting the desired amount of probe into the hybridization tube at 42°C, the tube is incubated further.
- Afterwards, the membrane is rinsed with a wash solution. On the other hand, autoradiography is used to observe the membrane.
Northern Blot Result Interpretation
Under radiography, the RNA bands can be seen as bands. You can measure RNA fragment length and semi-quantification by measuring the distance between bands and markers on your gel.
Uses of Northern Blot
- To identify and separate RNA fragments from diverse biological sources, the method can be utilised.
- A sensitive assay, Northern Blotting is used to identify transcription of DNA fragments that will be utilised as a probe in Southern Blotting.
- In addition, it enables for the identification and quantification of specific mRNAs in various tissues and live creatures.
- Northern blotting is utilised for gene expression investigations related to cancer-causing genes and gene expression after transplant rejects, among other applications.
- A molecular diagnostic test called Northern Blotting has been used to diagnose disorders including Crohn’s disease.
- The procedure is utilised to detect viral microRNAs, which play a key role in viral infection and transmission.
Northern Blot limitations
- Contrary to other current techniques, such as RT-PCR and nuclease protection tests, Northern blotting has a lesser sensitivity.
- For the procedure to work, a large volume of high-quality sample RNA must be obtained from the subject.
- Time-consuming and difficult, especially when many probes are required.
Northern Blot Citations
- Josefsen K, Nielsen H. Northern blotting analysis. Methods Mol Biol. 2011;703:87-105. doi: 10.1007/978-1-59745-248-9_7. PMID: 21125485.
- Blevins T. Northern blotting techniques for small RNAs. Methods Mol Biol. 2010;631:87-107. doi: 10.1007/978-1-60761-646-7_9. PMID: 20204871.
- Mishima, Eikan et al. “Immuno-Northern Blotting: Detection of RNA Modifications by Using Antibodies against Modified Nucleosides.” PloS one 10,11 e0143756. 25 Nov. 2015, doi:10.1371/journal.pone.0143756
- Koscianska, Edyta et al. “Northern blotting analysis of microRNAs, their precursors and RNA interference triggers.” BMC molecular biology 12 14. 11 Apr. 2011, doi:10.1186/1471-2199-12-14
- Brown T, Mackey K. Analysis of RNA by northern blot hybridization. Curr Protoc Hum Genet. 2001 Nov;Appendix 3:Appendix 3K. doi: 10.1002/0471142905.hga03ks30. PMID: 18428227.
Related Posts
- Phylum Porifera: Classification, Characteristics, Examples
- Dissecting Microscope (Stereo Microscope) Definition, Principle, Uses, Parts
- Epithelial Tissue Vs Connective Tissue: Definition, 16+ Differences, Examples
- 29+ Differences Between Arteries and Veins
- 31+ Differences Between DNA and RNA (DNA vs RNA)
- Eukaryotic Cells: Definition, Parts, Structure, Examples
- Centrifugal Force: Definition, Principle, Formula, Examples
- Asexual Vs Sexual Reproduction: Overview, 18+ Differences, Examples
- Glandular Epithelium: Location, Structure, Functions, Examples
- 25+ Differences between Invertebrates and Vertebrates
- Lineweaver–Burk Plot
- Cilia and Flagella: Definition, Structure, Functions and Diagram
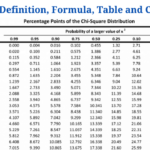
- P-value: Definition, Formula, Table and Calculation
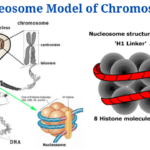
- Nucleosome Model of Chromosome
- Northern Blot: Overview, Principle, Procedure and Results