Nucleosome Model of Chromosome
- Chrosomes are self-replicating nucleic acid molecules that have particular organisational and functional characteristics and are essential for inheritance, mutation, variety and evolutionary advancement.
- Every chromosome’s structure is supported by many layers of DNA wrapped around proteins.
- Histones and non-histone chromosomal proteins are the traditional classifications for proteins which attach to DNA to construct eukaryotic chromosomes.
- Chromatin is the name given to the complex of both classes of proteins with the nuclear DNA of eukaryotic cells. Nuclear components, chromosomes are the building blocks of a specific organisation.
- Chromatin is a highly compacted structure made up of packed DNA that is required for DNA to fit into the nucleus.
- A succession of steps is required to assemble DNA into chromatin, starting with the synthesis of the nucleosome, and ending with a complex organisation of domains within the nucleus.
- DNA is condensed into an 11 nm fibre in the first step of this process, which indicates a 6–fold amount of compaction. Nucleosome assembly is used to do this.
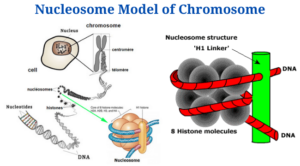
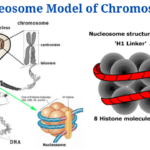
- The nucleosome is chromatin’s smallest structural component, formed by interactions between DNA and histone proteins.⦁
- It consists of an octamer core of histones H2A, B, 3 and 4 (or other histone variants in rare instances) and a DNA segment which surrounds the core of the nucleosome. Connecting nucleosome pairs is done with the help of “linker DNA”.
Introduction
- When DNA and related proteins are arranged in the nucleus, the nucleosome model helps explain how DNA and related proteins are arranged in chromosomes.
- Roger Kornberg proposed the model in 1974, and it is widely recognised model of chromatin organisation.
- It was confirmed and christened by P. Oudet et al., and it is the most widely accepted model of chromatin organisation (1975).
Features of the Nucleosome Model of Chromosomes
- DNA is securely bonded to an equivalent mass of histones in eukaryotes, forming nucleosomes, a repeating array of DNA-protein particles.
- Every human chromosome’s DNA double-helix will stretch thousands of times across the cell nucleus.
- Histones serve a critical funstion in packing this very long DNA molecule (i.e., nucleosome) into a nucleus with a diameter of only a few micrometres.
- Thus, nucleosomes are the chromatin’s primary packing unit particles, giving it a “beads-on-a-string” look in electron micrographs following treatments which unravel higher-order packing.
- In addition, every nucleosome comprises two copies of every one of the four nucleosome histones (H2A, H2B, H3, and H4), with a diameter of 11 nm and a height of 5.7 nm.
- Hexamer histones (H3, H4, H2A, and H2B) form the nucleus of the DNA helix, that is twisted 146 times around the histone octamer’s protein core.
- Every nucleosome is separated from the next by a 54-base pair stretch of linker DNA consisting of a single H1 histone molecule in chromatin.
- H1 molecules seal off two complete rotations around the histone octamers, which are 200 bp long.
- Nucleosomes repeat at 200 nucleotide or base pair intervals on average. For example, every human cell with 6 × 109 DNA nucleotide pairs includes
- 3 x 107 nucleosomes, and a eukaryotic gene of 10,000 nucleotide pairs will be connected with 50 nucleosomes.
Folding of DNA
- The DNA is initially assembled with a freshly synthesised tetramer (H3-H4), which is then selectively changed (e.g., H4 is acetylated at Lys5 and Lysl2 (H3-H4)) to produce a sub-nucleosomal particle, that is then followed by the addition of two H2A-H2B dimers.
- A nucleosomal core particle with 146 base pairs of DNA binds around the histone octamer as a result of this process. The nucleosome is made up of this core particle and its linker DNA.
- The maturation process requires ATP to ensure that the nucleosome cores are spaced evenly to create the nucleo-filament.
- The newly integrated histones are de-acetylated during this stage.
- Following the insertion of linker histones, the nucleo-filament is folded into the 30 nm fibre, the structure of which is still unknown.
- The solenoid model and the zig-zag model are the two main models.
- Finally, the nucleus develops a high level of organisation and particular domains as a result of further folding events.
Nucleosome Model of Chromosome Citations
- Verma, P. S., & Agrawal, V. K. (2006). Cell Biology, Genetics, Molecular Biology, Evolution & Ecology (1 ed.). S .Chand and company Ltd.
- Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K., & Walter, P. (2002). Molecular biology of the cell. New York: Garland Science.
- https://www.easybiologyclass.com/nucleosome-model-of-chromosomes-in-eukaryotes-short-notes/
- http://www.biologydiscussion.com/cell-biology/nucleosome-model/nucleosome-model-of-chromatin-assembly-cell-nucleus-biology/78886
- https://www.mechanobio.info/genome-regulation/what-are-nucleosomes/
- https://www.ncbi.nlm.nih.gov/pubmed/958895
Related Posts
- Phylum Porifera: Classification, Characteristics, Examples
- Dissecting Microscope (Stereo Microscope) Definition, Principle, Uses, Parts
- Epithelial Tissue Vs Connective Tissue: Definition, 16+ Differences, Examples
- 29+ Differences Between Arteries and Veins
- 31+ Differences Between DNA and RNA (DNA vs RNA)
- Eukaryotic Cells: Definition, Parts, Structure, Examples
- Centrifugal Force: Definition, Principle, Formula, Examples
- Asexual Vs Sexual Reproduction: Overview, 18+ Differences, Examples
- Glandular Epithelium: Location, Structure, Functions, Examples
- 25+ Differences between Invertebrates and Vertebrates
- Lineweaver–Burk Plot
- Cilia and Flagella: Definition, Structure, Functions and Diagram
- P-value: Definition, Formula, Table and Calculation
- Nucleosome Model of Chromosome
- Northern Blot: Overview, Principle, Procedure and Results